library(vegan)
pca_ord <- prcomp(USArrests, scale. = TRUE)
biplot(pca_ord)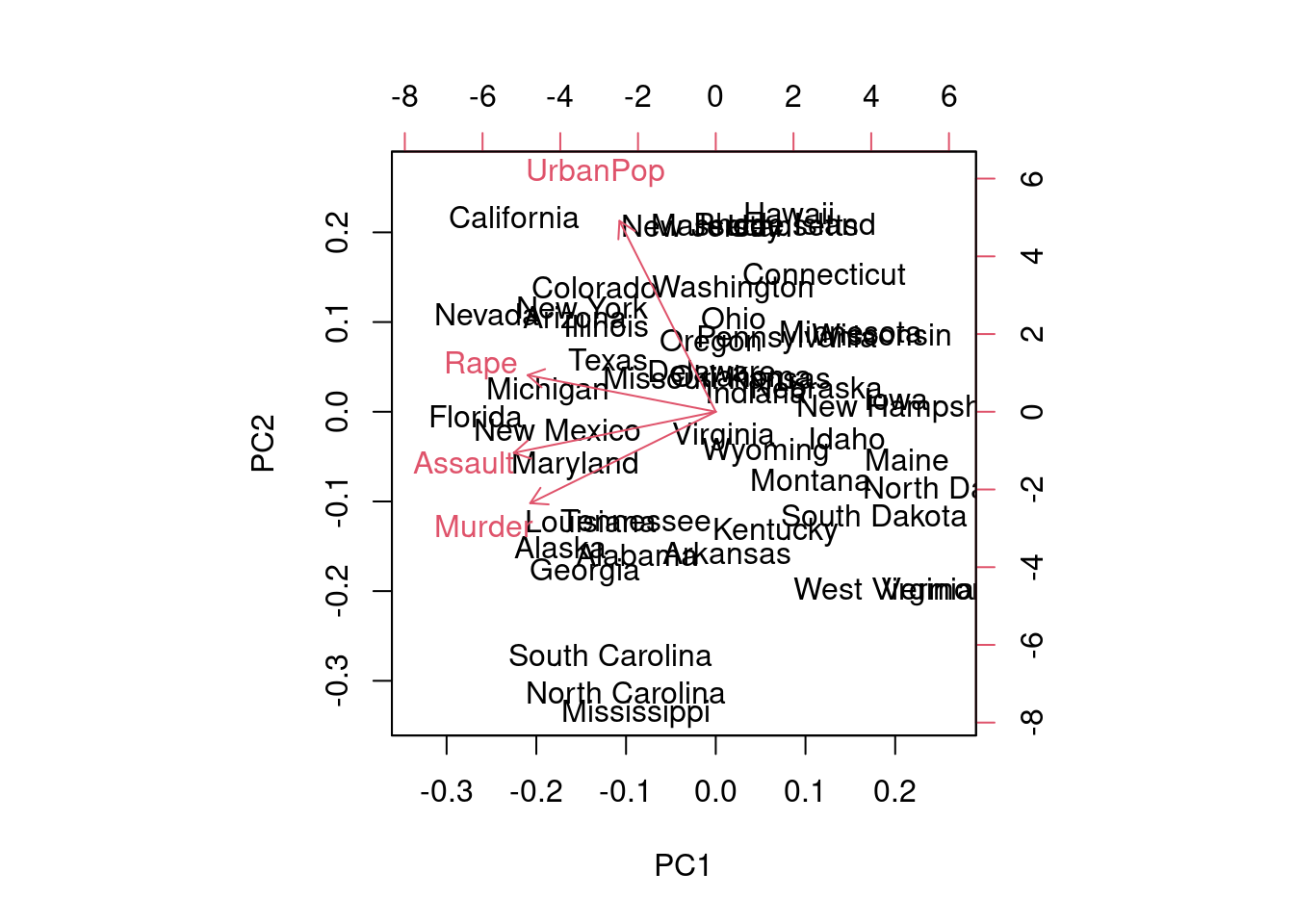
January 26, 2024
January 25, 2024
Principal component analysis (PCA) is used to reduce dimensionality in multivariate data analysis. This method is widely used in exploratory analysis and is better suited to the assessment of environmental variables. The primary objective of PCA is to transform the original variables into a new set of uncorrelated variables, called principal components, that capture the maximum variance in the data. These components are linear combinations of the original variables and are ranked according to the amount of variance they explain (eigenvalue). This method requires a rescaling of the variables (centering and standardisation) for a proper evaluation of the results.
In community analysis, PCA is better suited to assessing homogeneous vegetation and is prone to double-zero effects (inflated importance of rare species). In this respect, it is recommended to use transformation-based PCA instead.
Ordination applied to the wetlands datase.
stats::prcomp()vegan::rda() if only one table is provided as inputade4::dudi.pca()